To find the FAQs you are searching for, use the menu "plus buttons" below to open to the right category.
I can’t find the certificate of analysis for my product.
Please use the product number and batch number on the packaging box to identify the CoA or Technical Product Report (TPR) online. If you are unable to find it online, please send a request to cdx-technicalsupport@lgcgroup.com for assistance.
Where do I find the SDS for my Seraseq® product?
The Seraseq FFPE products have Safety Data Sheet (SDS) documents, which are available online on the product webpages; others don’t require an SDS document as those products don’t contain any substances which, at their given concentrations, are considered to be hazardous to health or the environment per GHS Classification in accordance with 29 CFR 1910 (OSHA HCS) and CLP Regulation (EC) No 1272/2008. Therefore, an SDS document has not been created for those products. A non-requirement letter can be provided in this case if needed.
What buffer is used for the purified DNA/RNA Mix format products?
Seraseq DNA Mix, RNA Mix and ctDNA Mix format products are provided in a 1 mM Tris, 0.1 mM EDTA, 10 mM NaCl, pH 8.0 aqueous buffer.
Is there a BED file (.BED) or vcf (.vcf) file available for each product?
Please contact cdx-technicalsupport@lgcgroup.com for assistance. VCF files are available for some but not all products.
Can I use any library kit with these reference materials?
Our products are designed to perform in the same way as patient samples. We and our customers have successfully used a variety of different library kits and sequencing platforms. In general, we recommend that you follow the manufacturer's protocol in using any kits or equipment.
Why are both the batch number and catalog number on the box different from those on the tube label? Are these supposed to be 2 different products?
For some of our products, the Material and lot numbers on the box are equivalent to those for a kit, whereas those on the tube labels are equivalent to part numbers. Both of these refer to the same product.
As the Certificates of Analysis always use the kit part and lot number, please refer to the lot number on the box when searching for or requesting documentation.
How are the multiplexed Seraseq reference materials made?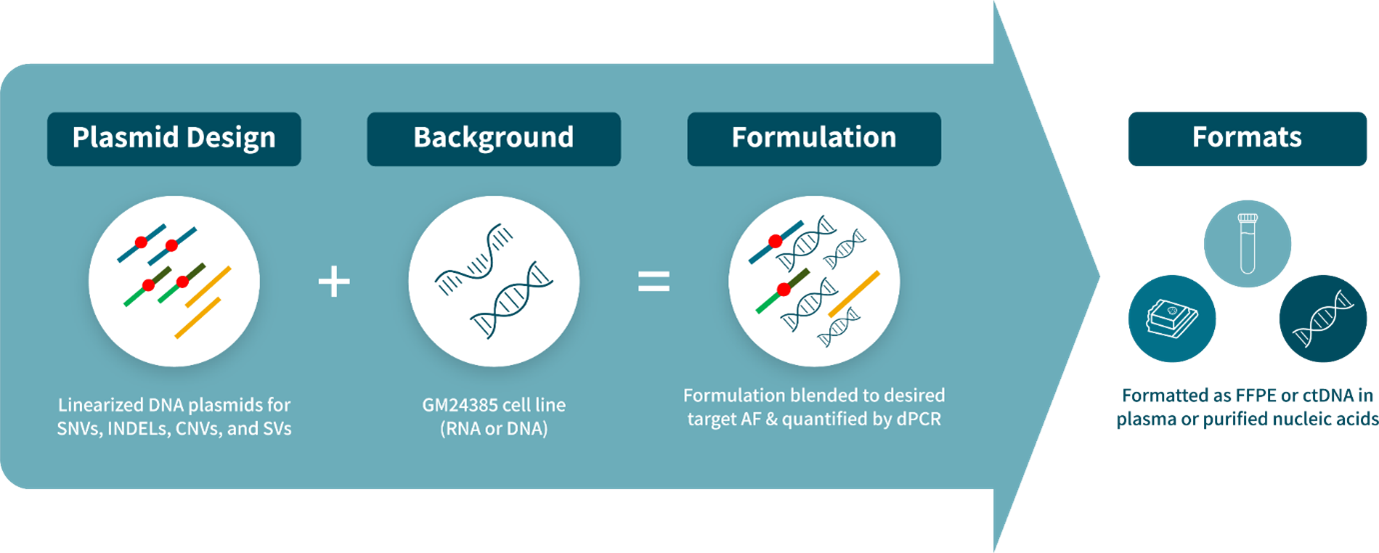 The Seraseq reference materials are based on biosynthetic DNA constructs bearing the variant of interest blended into a genomic DNA or total RNA wild-type or tumor background at the desired allele frequency or copy number, which is confirmed by digital PCR.
The Seraseq reference materials are based on biosynthetic DNA constructs bearing the variant of interest blended into a genomic DNA or total RNA wild-type or tumor background at the desired allele frequency or copy number, which is confirmed by digital PCR.
How do you quantitate the allelic frequencies of the introduced variants?
The quality control of allelic frequencies is performed using digital droplet PCR. The results are provided in the Product Report for each lot, which is available online. Depending on the product, the allelic frequencies are usually orthogonally confirmed with an appropriate targeted NGS assay.
How should I use Seraseq samples in my assay?
Seraseq samples should be treated in the same way as regular clinical specimens. The Mutation Mix format products can be introduced directly into the NGS library prep just like extracted DNA or RNA, while the Reference Material format samples need to undergo extraction first. Please see the product package insert for further details.
What is the difference between a Mutation Mix and a Reference Material?
Mutation Mix and Reference Material are two different sample formats. A mutation Mix is provided as purified DNA, ctDNA or RNA, whereas a Reference Material is a full-process sample provided either in FFPE or plasma, allowing the evaluation of the complete workflow from sample purification to result.
Is it possible to purchase just the background genomic DNA or total RNA from the GM24385 cell line?
Yes. We have 3 products, each containing a mixture of both DNA and RNA from GM24385, in either purified total nucleic acid or FFPE formats:
- Material number 0710-1580: Seraseq® TNA (DNA/RNA) WT Mix
- Material number 0710-0137: Seraseq® FFPE WT (DNA/RNA) Reference Material
- Material number 0710-1710: Seraseq® Compromised FFPE WT (DNA/RNA) Reference Material
Would it be possible to get the sequence file for the background cell line GM24385?
The GM24385 cell line is designated as HG002 by the Genome in a Bottle project. The genomic profiling datasets for HG002 can be downloaded from the GIAB website: https://www.nist.gov/programs-projects/genome-bottle.
Where can I find the exact genomic variant coordinates or RNA fusions / translocation sequence?
We provide a Technical spreadsheet upon request. Please contact cdx-technicalsupport@lgcgroup.com for assistance.
Do the Fusion RNA Mix products contain any DNA ?
No. Our Fusion RNA products contain the RNA fusion fragments blended with total RNA from the wild type GM24385. They are treated with DNase and there is no DNA present.
Do the Fusion RNA FFPE Reference Materials contain any DNA ?
The normal DNA from the background cell line might be present depending on the RNA extraction protocol used, however the introduced fusion transcripts are only present on the RNA level and not as DNA translocations.
What yield should I expect from the FFPE DNA / FFPE RNA products?
The DNA or RNA yields of our FFPE products are listed in the Technical Product Report or Certificate of Analysis. Please refer to those documents for details. We cannot guarantee the same yield will be obtained using a different type of extraction kit than the one(s) listed. In general, you should expect at least 100 ng DNA or 400 ng RNA per FFPE curl when using the QIAamp DNA FFPE or Agencourt Formapure RNA kit, respectively.
How should I process the FFPE Reference Materials?
You should process the FFPE Reference Materials in the same way as you process patient samples in FFPE format. However, keep in mind that the Seraseq FFPE Reference Materials are derived from cell pellets, which might behave slightly differently from tissues when using mechanical force to break the cells. Thus, the extraction conditions might be adjusted accordingly. For example, the Seraseq FFPE curls are lighter and more transparent than clinical samples so the curl and resulting pellet are smaller and easier to lose during the purification protocol. The advice is to extract the FFPE reference material alongside a clinical sample, being careful with the handling to keep track of the curl / pellet location during the entire protocol. Similarly, optimization might be needed for automated extraction in order to achieve a good DNA or RNA yield.
Can I obtain the exact AF values for the Compromised FFPE Tumor DNA Reference Material?
Yes, please contact cdx-technicalsupport@lgcgroup.com.
Do the RNA fusion construct have a polyA tail?
Yes, all synthetic fusion transcripts include a polyA tail and can be used in any cDNA library construction protocols.
How should I analyze the genomic DNA and FFPE TMB reference materials?
TMB describes the number of mutations per megabase of genome, which is best measured using whole exome sequencing (WES), while targeted panels with large enough coverage have also been shown to generate comparable TMB scores. The TMB scores of our tissue TMB products have been evaluated using WES in a tumor-normal workflow following the Friends of Cancer Research recommendations. The sequencing and data analysis conditions are described in the Data Sheet and TPR and can be used to benchmark the results of your TMB assay. Please refer to those documents for details.
How should I analyze the bTMB reference materials?
As there is less consensus in the community around bTMB analytics, we are working with key opinion leaders and vendors to establish best practices. Until now, bTMB has been practical only using moderately-sized panels rather than WES. As a result, the parts of the genome covered by the panel and related analysis pipeline will vary depending on the panel used. The TMB scores of our bTMB products have been evaluated using the TruSight Oncology 500 panel with the default TMB analysis parameters. The sequencing and data analysis conditions are described in the Data Sheet and TPR and can be used to benchmark the results of your TMB assay. Please refer to those documents for details.
What is the difference between the MSI Reference Panels and MSI-H? When should I use which product?
The Seraseq MSI Reference Panels contain biosynthetic spike-ins of 5 clinically approved MSI loci typically analyzed in qPCR/NGS assays at AF5% or AF20%, confirmed by dPCR. They are provided as a tumor-normal matched set and are suitable for qPCR, qPCR/CE and NGS analysis analyzing these defined loci only.
The Seraseq gDNA MSI-High Mix and FFPE Reference Materials are based on a human tumor cell line harboring multiple MSI loci (including the 5 loci present in the MSI Panels) at close to 100% VAF and is provided in a tumor-only format. It has been tested by 2 orthogonal NGS assays and can be used as a positive MSI-H control for NGS assays analyzing the MSI status based on any number and type of genomic loci.
In what formats are the products available for sale?
Two formats – (1) Purified ctDNA (in a 1 mM Tris, 0.1 mM EDTA, 10 mM NaCl, pH 8.0 aqueous buffer) “Mutation Mix”, and (2) encapsulated format (plasma-like material) “Reference Material”.
What are the VAF ranges for the ctDNA products?
- Seraseq Myeloid ctDNA Mix: 0% (WT), 0.1%, 0.5%, 1.0%
- Seraseq ctDNA v2 Reference Material: 0% (WT), 0.125%. 0.5%, 1%, 2%
- Seraseq ctDNA v2 Mutation Mix: 0% (WT), 0.125%, 0.25%, 0.5%, 1%, 2%
- Seraseq Complete Reference Material: 0% (WT), 0.1%, 0.5%, 1%, 2.5%, 5%
- Seraseq Complete Mutation Mix: 0% (WT), 0.1%, 0.5%, 1%, 2.5%, 5%
How are these VAFs determined?
We use digital PCR assays on a BioRad QX200 for VAF quantification for each allelic blend. We also confirm the VAFs using the ArcherDx Reveal ctDNA 28 assay. Measured allele frequencies can be found in the individual Technical Product Reports or CoAs.
What NGS chemistry is recommended to analyze these reference samples?
The Seraseq ctDNA reference materials are designed for targeted NGS assays based on amplicon or hybrid-capture chemistries. These should be compatible with most common NGS assays from Illumina, ThermoFisher, Archer, Qiagen or Roche Dx. In-house, we use the ArcherDx® Reveal ctDNA® 28 kit on a MiSeq v2 chemistry for NGS analysis of the ctDNA reference materials. Data analysis is performed using the ArcherDx bioinformatics pipeline. The data VCF and BED files are available to customers upon request.
What is the recommended ctDNA reference sample input for NGS assay analysis?
We do not recommend any specific ctDNA input for NGS analysis. Our materials perform similarly to patient samples with regards to library incorporation in most assays.
How do I use these reference samples in my NGS workflow?
The Mutation Mix format is ready to use in a sequencing library enrichment assay (amplicon- or hybrid capture-based) following the assay vendor protocol for NGS analysis of circulating cell-free DNA samples, or protocol developed in your laboratory. No further DNA purification is required. The encapsulated (plasma-like) reference samples may be used in a manner consistent with plasma fractions prior to extraction, in parallel with your test specimens in a target enrichment NGS workflow following the protocol as described by the assay vendor.
For the encapsulated format, does SeraCare recommend a cfDNA extraction kit?
No, we do not officially recommend an extraction kit for use with the Seraseq ctDNA reference materials. However, we provide information in our Package Insert showing our results using the QIAGEN QIAamp® Circulating Nucleic Acid Kit for ctDNA extraction, and the Qubit dsDNA BR Assay Kit to quantitate the yield. More details on this are provided in the product Package Insert.
What is the typical yield of ctDNA recovered from a single vial of the RM product?
Using the cfDNA extraction kit from Qiagen and Qubit for quantitation, typical ctDNA yields from a 1 mL volume of encapsulated material range between ~25-35 ng/mL This is based on triplicate extractions.
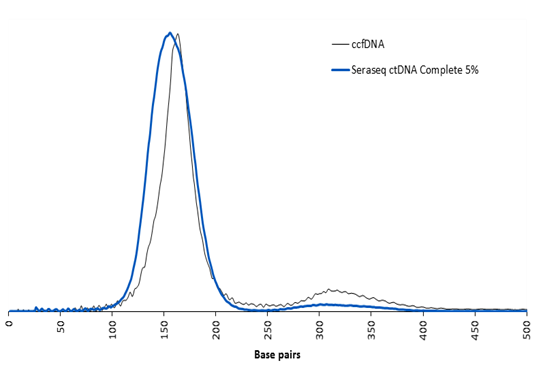
What is the typical ctDNA fragment size in the Seraseq ctDNA products?
Typical range is 155-185 bp across all VAFs and WT samples.
An example for the ctDNA fragment size, as determined using the Agilent Bioanalyzer is shown below for the ctDNA Complete™ AF5% versus natural circulating cell free DNA from a patient plasma sample.
What is an appropriate LoD for my ctDNA assay?
LoD will vary with the assay and sequencing platform used. Many assays seem to perform well down to the 1-0.5% VAF range, but this can be empirically determined with our reference materials. We do not provide technical support for issues arising from end-user dilution of VAF, thus we recommend using our material to validate LoD >0.1%.
Is the background free of mutations?
Our ctDNA materials are generated using GM24385 cells. The propagation and production of our materials maintains a background that is generally consistent with the published genome sequence of GM24385. That said, at VAF <1% there may be artifacts from the production of the products. Our materials are meant to be used as a sensitivity control to verify the ability to measure the VAFs and CNVs stated in the product report for a given batch of material.
Are there any assays that do not work with SeraCare ctDNA materials?
All commercially released assays should be compatible with SeraCare ctDNA material. That said, we have not tested every assay on the market. If your assays are sensitive to residual biotin, please purify the SeraCare ctDNA materials using streptavidin beads before using them.
Can you clarify the descriptions of CNVs on the ctDNA Complete product reports?
The additional copy numbers of our ctDNA Complete products were designed to be a similar order of magnitude as the AF. For example, AF 1% will have roughly +1 apparent copy of the CNV genes, AF 5% will have roughly +5 apparent copies of the CNV genes. The equivalent additional copy numbers of the CNV genes in tumor cells are: the numbers of the additional copy X 100 / (2 X AF%).
Which ctDNA offering is best for my assay?
We generally advise our customers to look at the VAF ranges and variant lists to guide the decision between ctDNA v2 and ctDNA Complete. If both options seem sufficient for the needs of your laboratory, ctDNA Complete is a newer product and has some minor improvements to its production process.
How do I use the MRD ctDNA Panel?
The MRD ctDNA Panel mix is designed to support development and validation of ctDNA-based patient-informed minimal residual disease (MRD) assays, mainly for solid tumors. It is based on a human tumor cell line harboring a high number of variants, which is analyzed by WES and blended to different tumor fractions with its matched normal and clinically actionable biosynthetic variant spike-ins. This allows the user to design a sample-specific MRD assay targeting any of the variants known to be present and validate the assay sensitivity and performance at the different tumor fractions.
How are the VAFs in the MRD ctDNA Panel tested?
The VAFs of the spiked-in variants are checked by a commercial targeted NGS panel at the 0.25% VAF (target frequency in the 0.5% tumor fraction sample). All tumor fractions are analyzed by a custom targeted NGS panel in each tumor fraction dilution and the total number of variants detected are compared with a theoretical statistical model to confirm the expected dilution behaviour. Please see the MRD data sheet and white paper [LINKS] for more information.
How do you generate multiplexed reference samples with a single genetic background?
The Seraseq inherited disease reference materials are based on biosynthetic DNA constructs bearing variants of interest blended into a wild-type genomic DNA background at the desired allele frequency or copy number, which is confirmed by digital PCR.
How do you quantitate the allelic frequencies of the introduced variants?
The quality control of allelic frequencies is performed using digital droplet PCR. The results are provided in the Product Report, which is available online.
How do I use these materials with my assay?
Seraseq inherited disease samples are provided as purified high molecular weight genomic DNA, and can enter the workflow as a regular specimen, after extraction. Please see the package insert for further details.
What NIPT reference materials are available?
The Seraseq® portfolio of matched maternal-fetal NIPT reference materials includes the most frequent chromosomal aneuploidies (trisomy 21, 18 and 13), sex chromosome aneuploidies, 22q11 microdeletion (associated with DiGeorge Syndrome), as well as euploid reference materials to be used as negative controls.
These materials are purpose-built to enable monitoring of the full NIPT process from extraction through the reporting of assay results. Refer to our product list for more information.
How are these products made?
Thanks to an exclusive collaboration with Stanford Medicine and Institutional Review Board (IRB) approval, we are able to acquire rare patient donors’ blood samples whose pregnancies have been complicated by a genetic abnormality.
Seraseq maternal-fetal matched (i.e., related) reference materials are developed using a proprietary technology. Cell-free DNA (cfDNA) is extracted from the plasma of the pregnant patient carrying either a euploid (normal) pregnancy or one with a confirmed aneuploidy or microdeletion.
The next step is the amplification of the cfDNA such that the natural fetal and maternal cfDNA size profiles, as well as all of the DNA characteristics of the original clinical sample (fetal fraction, SNP and GC content, etc.) are maintained. This DNA is then stabilized via encapsulation into liposomes, purified, and introduced into SeraCon™ Matribase simulated plasma matrix.
What is SeraCon Matribase?
SeraCon Matribase is a defibrinated plasma product manufactured in our ISO13485 registered facility, with characteristics similar to human plasma. It is sourced from plasma collected from FDA-licensed blood centers, with testing carried out at the donor level, that is fully traceable through the manufacturing process. With preservatives and stabilizers, Matribase is the ideal source of stable plasma to use as a diluent.
What is the fetal fraction of Seraseq Matched NIPT reference materials and how is it estimated?
Our materials’ fetal fractions vary depending on the source patient sample. The fetal fraction is assessed by an external NIPT laboratory before being released; this information is provided in the Technical Product Report (TPR). However, it is important to keep in mind that fetal fraction assessment varies significantly among commercially available NIPT assays. Some of the fetal fraction estimation methods include:
- Statistical computation of sequence counts.
- Y-sequence (male fetus only).
- Fragment length ratio.
- SNP patterns.
The information provided in the TPR should be used for guidance only. Fetal fraction measurements should be empirically determined for NIPT methods other than those indicated in the TPR.
Is the fetal fraction customizable?
Yes. For some of the source samples, we can dilute down the fetal fraction of the resulting Seraseq reference materials and obtain a range of fetal fractions to test the limit of detection of an assay. Please refer to our custom services page to obtain more information.
How should I use reference materials in my assay?
Seraseq NIPT reference materials should be processed in the same way as patient samples, following the protocol as described by the assay vendor. They are encapsulated (plasma-like) cfDNA samples, which may be used in a manner consistent with clinical plasma samples starting with cfDNA extraction. Test results show that Seraseq NIPT reference materials behave closely to real-world clinical samples, regardless of the NIPT platform and method used, such as whole genome or targeted NGS or digital PCR.
Can I dilute Seraseq NIPT reference materials?
Each vial is formulated as a 1 mL plasma-like sample containing approximately the same amount of cfDNA as 4 mL of plasma obtained from a patient. Note: depending on cfDNA extraction methodology, recovery yields may vary.
Different NIPT assays use different starting volumes of plasma (between 1 mL and 4 mL). It is possible to dilute our reference materials to obtain the minimum volumes required for your workflow. The most common diluents for Seraseq NIPT reference materials are PBS and SeraCon Matribase. Unprocessed patient plasma, either commercially sourced or otherwise, should not be used due to the presence of naturally-occurring cfDNA, which will cause aberrant results. This dilution should be done immediately before use. We do not recommended putting the reference materials in Streck tubes or any tubes containing a fixing agent, only tubes compatible with the platform workflow should be used.
What is the cfDNA recovery after extraction?
There is variability between the methods used and between the labs using the same method. It is a good idea to determine what percent recovery is normal for your method and NIPT assay. Our Technical Product Report (TPR) describes yield per sample for a single extraction and quantification scenario. Measured yields may vary between methods, but our materials have been designed and verified to be compatible with most commercially-available NIPT assays.
What are the advantages of Seraseq Matched NIPT reference materials?
Matched maternal-fetal reference materials derived from pregnant maternal plasma preserve the SNP content of mother and fetus, as well as the size difference between the fetal and maternal cfDNA. They enable accurate assessment of a broad range of NIPT assays, including those based on counting methods, SNP analysis, and utilizing the fetal-maternal cfDNA size difference.
Seraseq reference materials are manufactured in GMP-compliant and ISO13485-certified facilities, ensuring lot-to-lot consistency. They are used globally by assay manufacturers, laboratories as well as proficiency providers.
Evaluating multiple methods using the same exact material provides an objective comparison of performance. Any changes in a workflow due to an upgrade of the method can also be easily compared by using Seraseq reference materials. Training new staff is very important and using reference materials makes the process easier and comparable not just among operators, but also over time.
Our reference materials are also useful for troubleshooting assays that are deployed across multiple sites. The long shelf-life of the materials for repeated use of the same lot for training, validation, or assay performance assessment.
What is the recommended storage temperature and expected shelf life for Seraseq NIPT reference materials?
Seraseq NIPT reference materials are formulated as stabilized encapsulated cfDNA in SeraCon Matribase simulated plasma. They are shipped on ice packs, with a recommended storage temperature of 2-8°C. and should not be frozen. When properly stored, the shelf life is 4 years from date of manufacture.
Test methods and techniques may produce variable results and are for informational use only; therefore, we recommended that customers evaluate the material for suitability and shelf life according to their intended uses.
How is the ploidy status verified?
Pregnant patients are referred following a high-risk NIPT and/or anomalies identified on their scan. Diagnostic testing such as karyotyping is then performed to confirm the ploidy status of the fetus and any other findings.
After manufacturing the reference materials from the patient sample, a further check is done by at least one external NIPT laboratory. Information on the specific NIPT assay used is indicated in the Technical Product Report (TPR) for each lot.
What is the difference between Matched Reference Materials and the previously available unmatched Aneuploidy Reference Materials?
Our second-generation maternal-fetal matched reference materials are made using pregnant patient blood samples and contain both maternal and fetal cfDNA while the first-generation (unmatched) aneuploidy reference materials were manufactured using a robust biosynthetic technology from unrelated maternal-fetal source materials.
First, male fetal DNA derived from placental trophoblast cells with a confirmed trisomy was blended with genomic DNA obtained from an unrelated, non-pregnant female to create predetermined fetal fractions. The DNA was then fragmented to mimic a cfDNA size distribution of around 170bp, followed by encapsulation in Matribase plasma matrix to create full-process controls. The main advantage of the unmatched materials is the precise control of fetal fraction. They can also perform better with some assays, such as the rolling-circle amplification-based Vanadis assay. Please note that the unmatched cell-line reference materials are only available on a custom request basis. Please contact us for more information.
What is the NIPT test compatibility of these products?
The Seraseq maternal-fetal matched NIPT Reference Materials have been tested with a broad range of chromosome counting and SNP-based NIPT methods, such as Illumina’s VeriSeq v2, Roche’s Harmony®, Natera’s Panorama™, Yourgene Health’s IONA® and Sage™ tests, as well as various Laboratory Developed Tests. These materials are not compatible with the PerkinElmer Vanadis® assay, for which our cell-line derived reference materials would be more appropriate. Please also note that these materials are not suitable for methylation-based assays.
Are these products customizable?
Yes. We offer various custom options with a high degree of flexibility regarding concentration, fetal fraction, format, volume or blending to create mosaic samples. By leveraging our extensive biobank of patient-derived cfDNA, we can offer reference materials for aneuploidies and microdeletions, Rhesus factor and other syndromes.



